Reduce the Runtime & Memory Requirements of your Workloads by more than 50% with Accelerated Datasets
Bioinformaticians and ML engineers often need to balance performance versus cost when running data-intensive workloads. Union optimizes for reducing cost by running ephemeral compute, which means reducing idle resources. This can lead to some runtime overhead when the data used by those resources scales up. To speed this up, we introduce Accelerated Datasets, a way of mounting and caching large datasets to be quickly consumed by your workflows.
Setting up the problem
Union workflows are containerized, meaning that each task (step) runs in its own container. This paradigm promotes reproducibility and rapid development while supporting highly complex workflows (DAGs) composed of heterogeneous steps. One drawback of containerization is the fact that each execution environment must be created before a task can run. In practice, a task that takes a large dataset as an input will download the data into ephemeral storage on a Kubernetes pod, then perform in-memory logic as specified in the user’s code. If this task is run one hundred times, data is downloaded one hundred times. Situations can arise where the same expensive operation is happening over and over for similar tasks.
With ephemeral compute, the underlying execution engine needs to:
- Spin up the compute resource (i.e. a VM and a pod to run the task)
- Download the data into the task pod
- Execute user code
- Move on to the next task (terminating the compute resource)
In situations where datasets are large and user code is relatively lightweight, the cost of setting up the execution environment can vastly exceed the actual execution time. This presents a high-impact opportunity to help users develop and run workflows more quickly by reducing the runtime associated with data downloads. It also directly reduces compute costs by having ephemeral resources around for less time.
Introducing Accelerated Datasets
In machine learning, model training can re-use the same dataset across multiple runs with different parameters. In data engineering, a large, raw dataset might be processed in multiple ways to fulfill downstream use cases like business analytics. A common use case in bioinformatics is to align DNA sample reads to a large, static reference genome. What if we could cache these files to a semi-persistent data store on the underlying node?
This is exactly what we did to help our customers save a huge amount of time, especially when working with homogenous workflows that produce wide map task fanouts. We’re calling this capability Accelerated Datasets. It allows you to pre-load static, read-only datasets into compute nodes and reuse them to reduce a major part of the overhead incurred by using ephemeral compute resources. Below we detail how DelveBio, one of our customers in the medical diagnostics space, found that Accelerated Datasets quadrupled their throughput on the same nodepool.
Implementation challenges
Implementing this feature required several considerations. High-throughput, persistent storage is naturally at odds with ephemeral compute. While there are obvious cost savings in scaling away idle nodes, striking a balance with performance around large datasets is essential to benefiting from that optimization.
We evaluated a few options at first:
- Stream assets in and consume the data incrementally. This may work for some use cases, but it doesn’t work if the workload relies on random access.
- Setting the node minimum to one and pulling in any static assets at provisioning time. This leads to a lot of idle compute, but can still be a good option if it’s consistently used at or close to capacity.
- Mounting an EBS disk as a persistent volume. This would work, but you get mediocre performance, especially for random reads and writes. This is because it’s essentially an NFS (network file system) share under the hood.
- Using S3fs-fuse as a file system in userspace. This works in the AWS cloud ecosystem, but we noticed similarly poor performance as well as being significantly more expensive than naive S3 fetches due to all the additional API calls.
The solution: S3 mountpoints
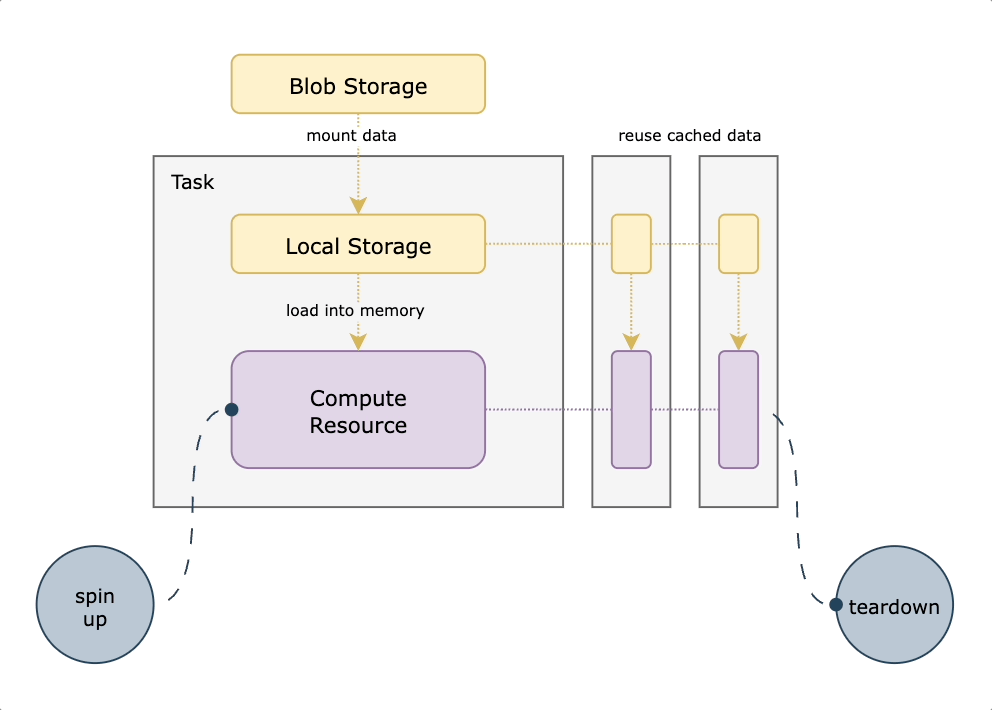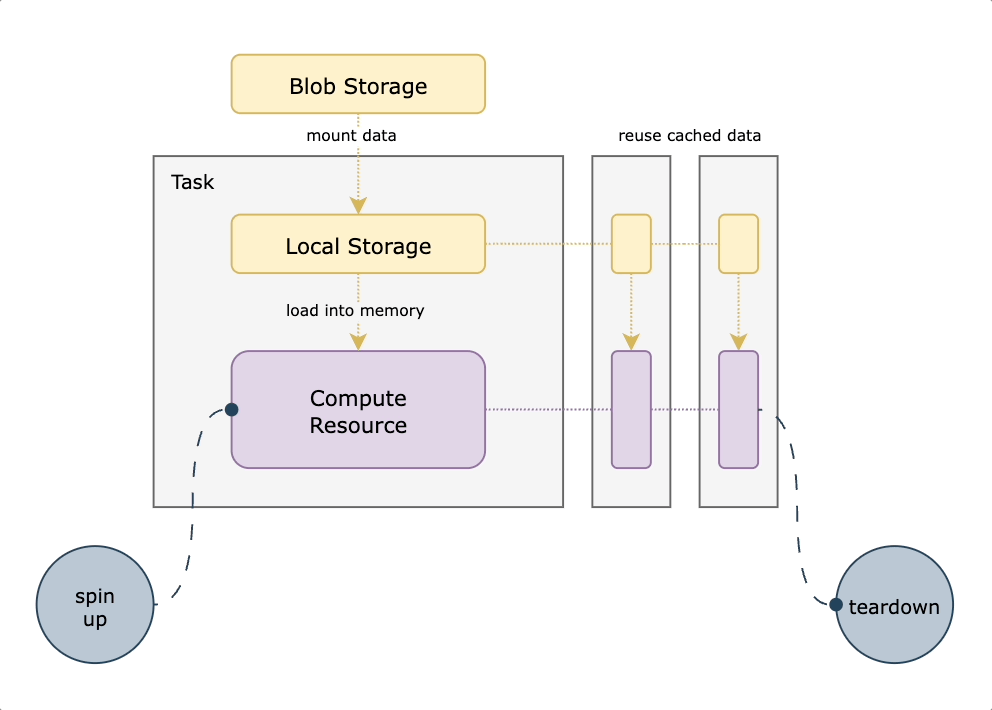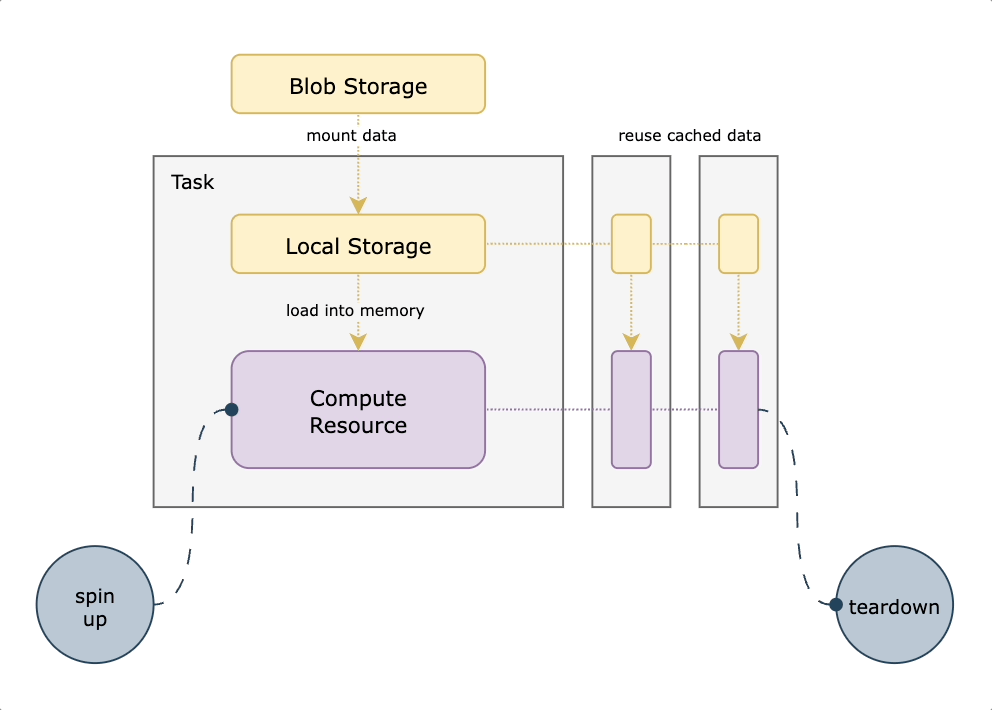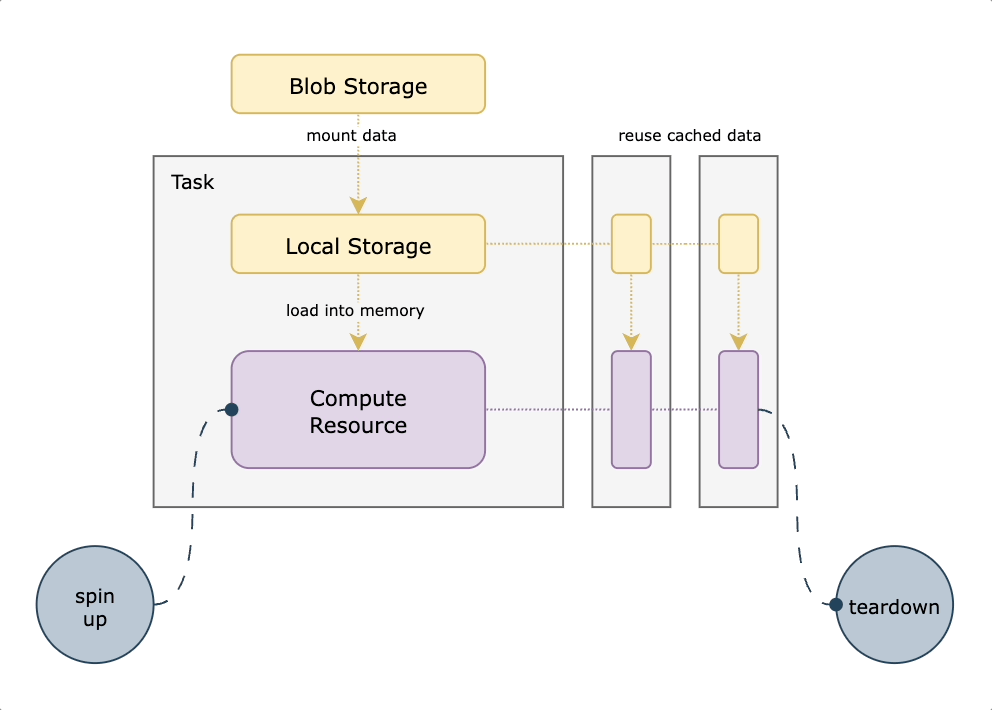
The initial implementation of this capability prefetched all the assets in the bucket during node-provisioning and stored them in a local persistent volume, which would then be mounted on the task pod at startup time. This led to great performance out of the gate, since all assets were already present locally. However, it required some client side code to ensure things had finished downloading. This meant that any tasks needing immediate access to the data upon node startup would have to wait until everything had finished downloading, leading to a poor developer experience and a higher lift from existing workflows.
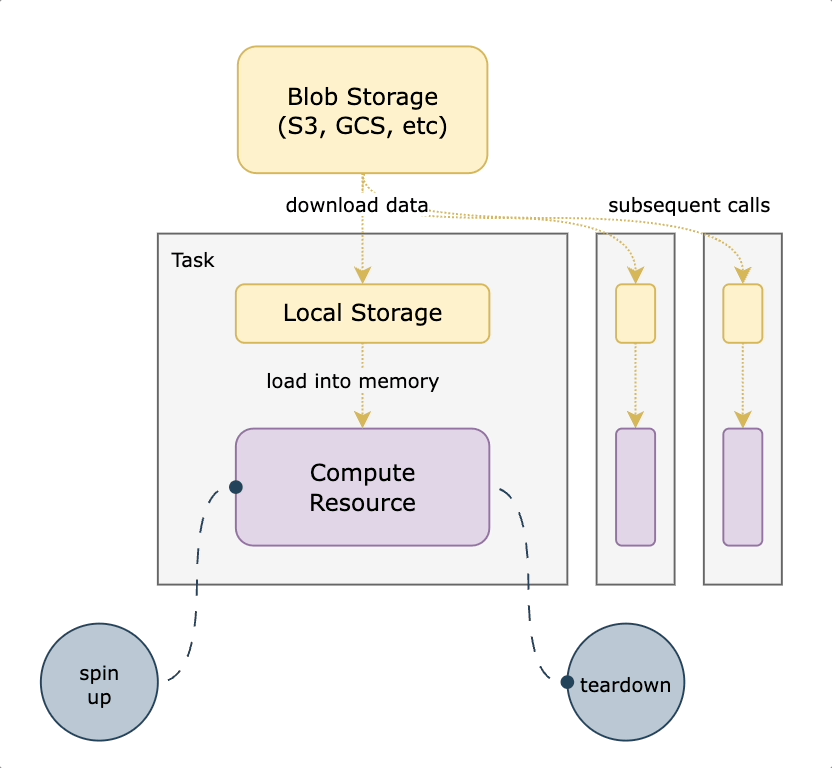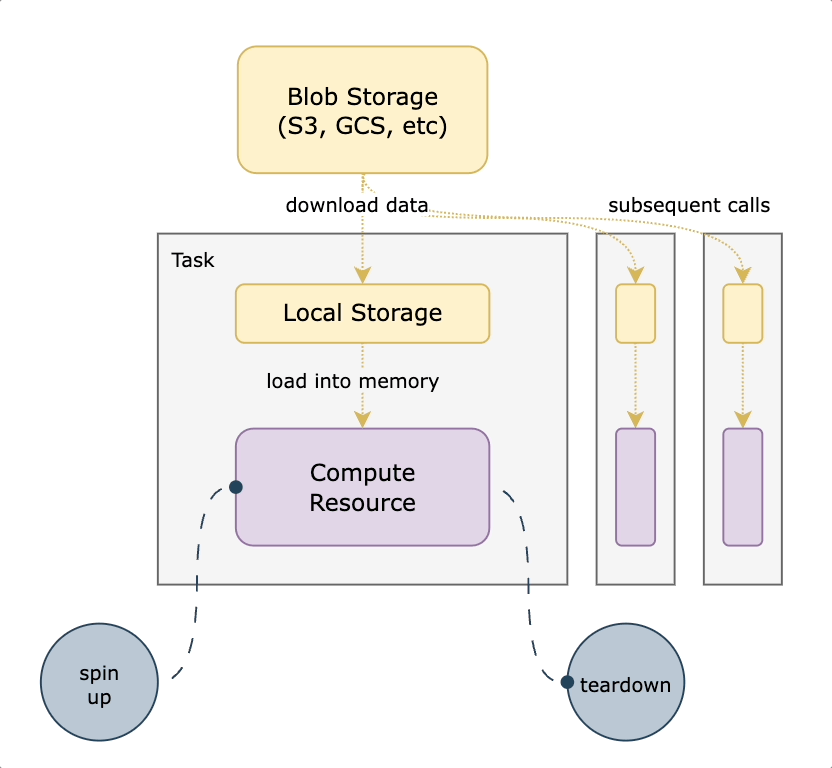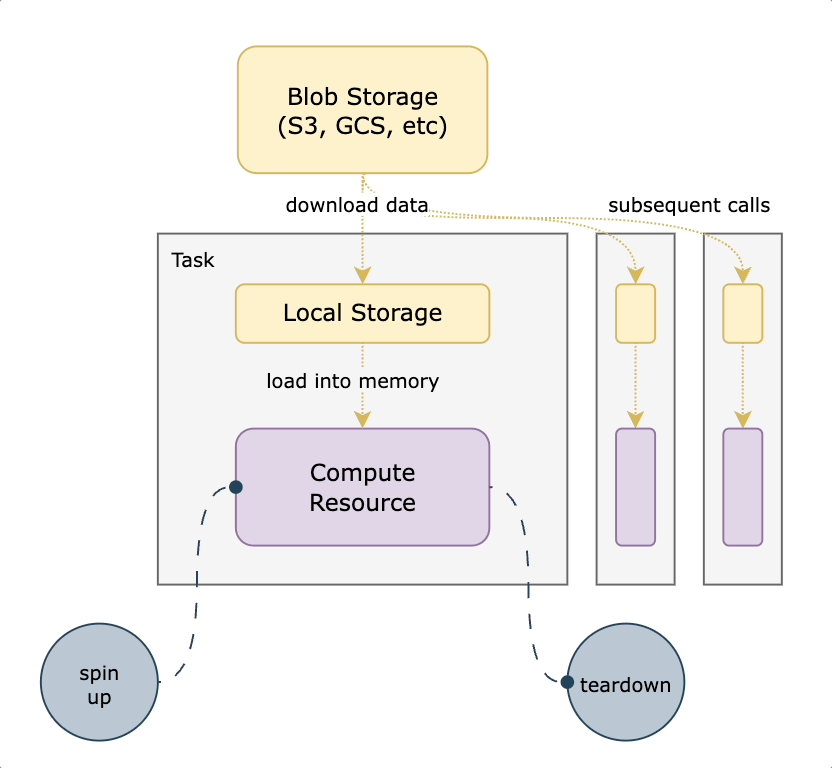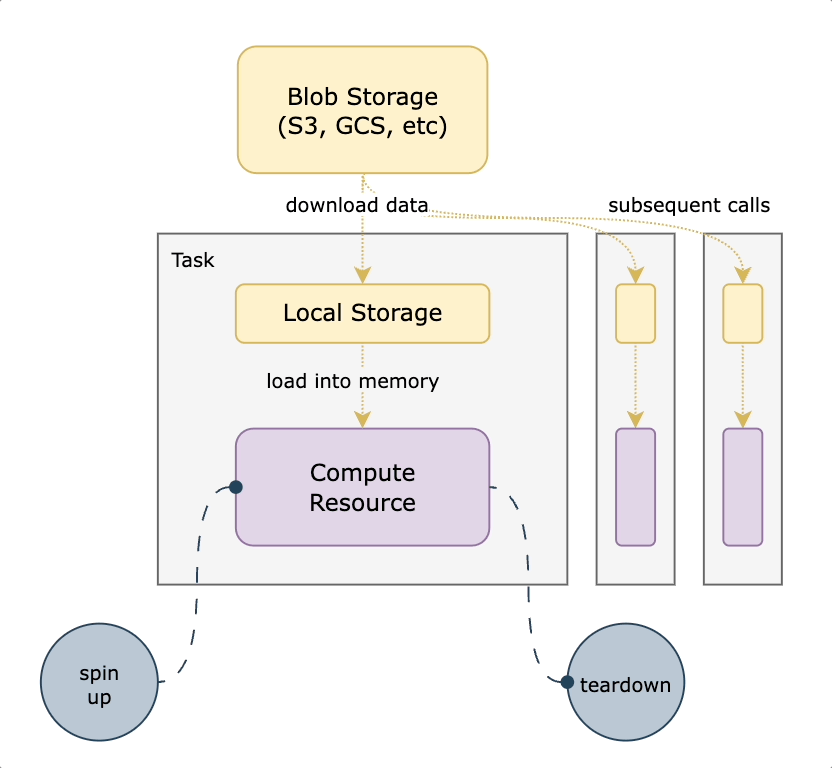
We decided to use S3 mountpoint. While similar to `S3fs-fuse` in that it translates filesystem calls to equivalent API operations, it benefits from caching and being a native AWS offering. With this approach, we mount a well-known persistent datasets bucket to all nodes on startup. We then translate any `FlyteFile` or `FlyteDirectory` objects originating from that bucket into their local filesystem equivalents, leading to a very small lift from non-accelerated tasks. An additional benefit of translating the URI to its native filesystem equivalent is the ability to do all standard Python path operations on the `FlyteFile.path`, since it’s a truly local object. This means you’re not reliant on which methods are implemented by FlyteFile/FlyteDirectory or having to call `download()` first. The diagram below shows how accelerated datasets make your tasks more efficient by reusing cached data mounted on the task pod.
A real-world example: genome sequence alignment
Here’s a real-world example of an alignment workflow across a number of different samples:
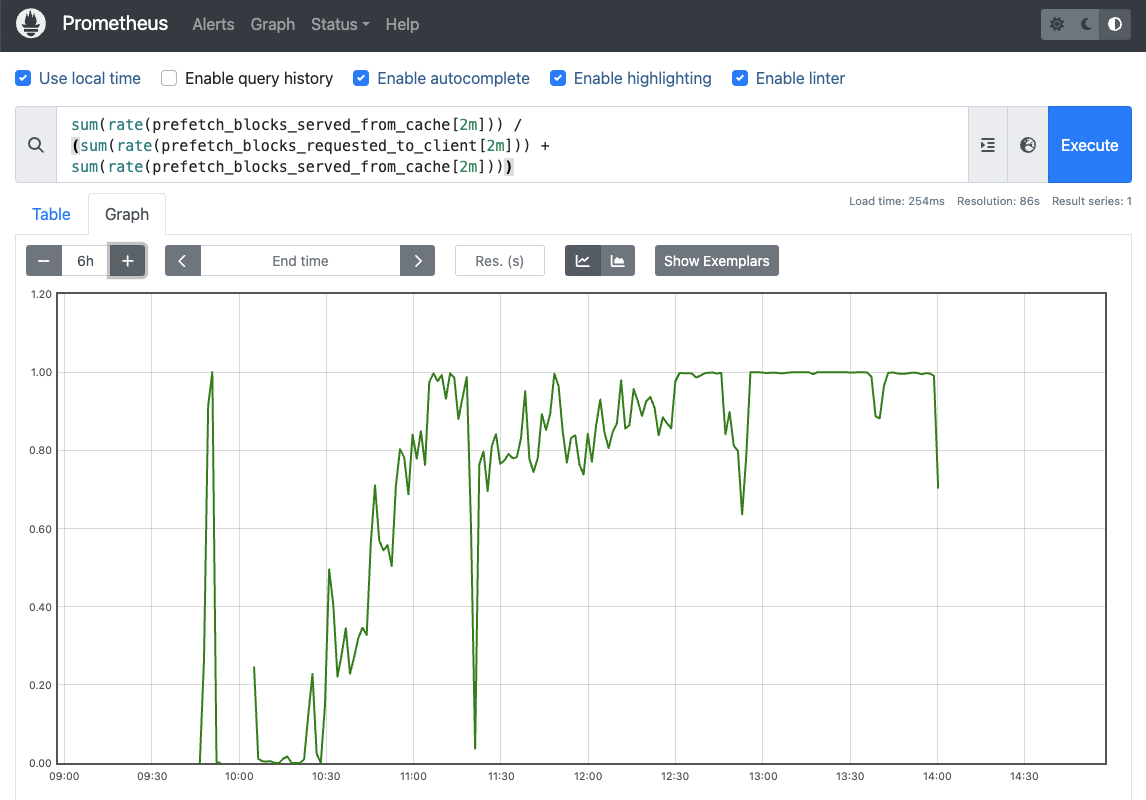
The index input `idx` being passed to the alignment task is the salient aspect here, located at `s3://unionai-persistent/bowtie2-index/`. If this were run on a single sample, the gains would be fairly unimpressive. However, the cache will become more populated as more samples are processed, which translates to higher hit-rates. This is captured below from another real-world dashboard showing the cache hit-rate climb and eventually level off for significant periods of time at 100%, showing that the asset is almost completely cached:
Benchmarking with a simple task
To illustrate the kinds of speedups that are possible over a naive fetch, we ran the task in the code snippet below thirty times in a dynamic workflow with `maxParallelism=1` so that we ensure that the same node is performing the task. It’s a simple MD5 sum operation and comparison with runtimes plotted at the end.
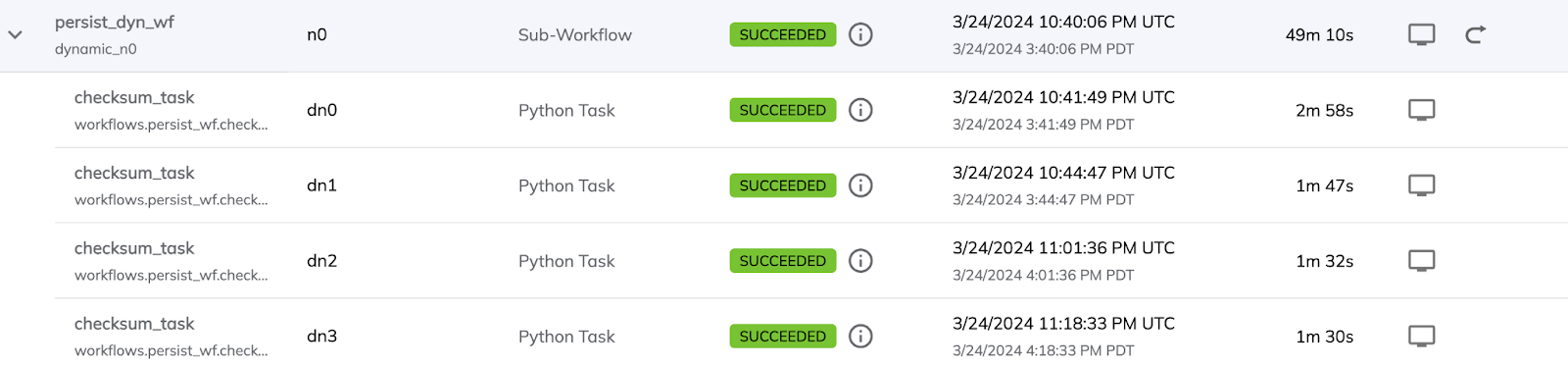
Screenshots from the Union UI show a total runtime of the dynamic workflow followed by a few of the benchmarking tasks.
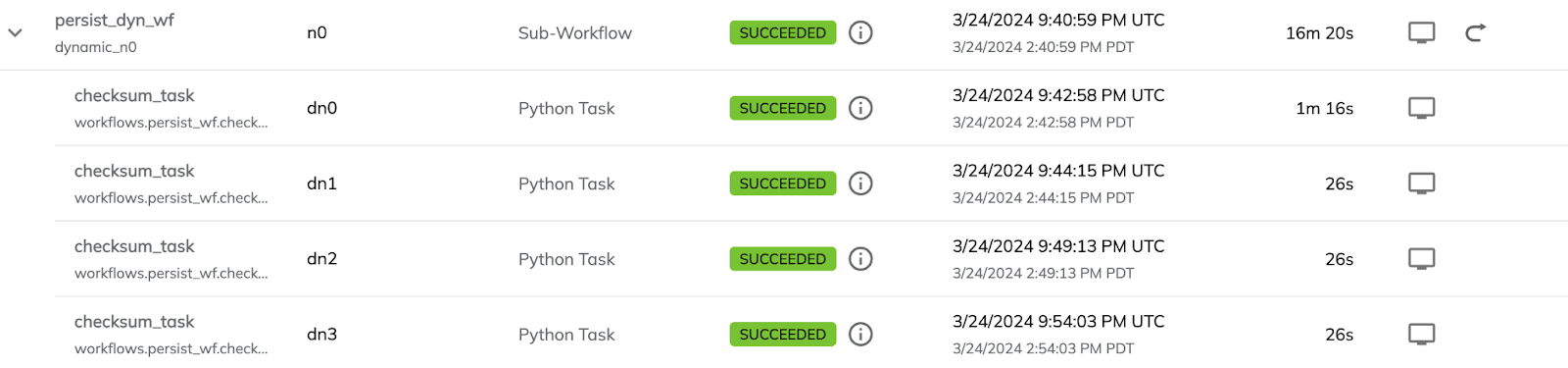
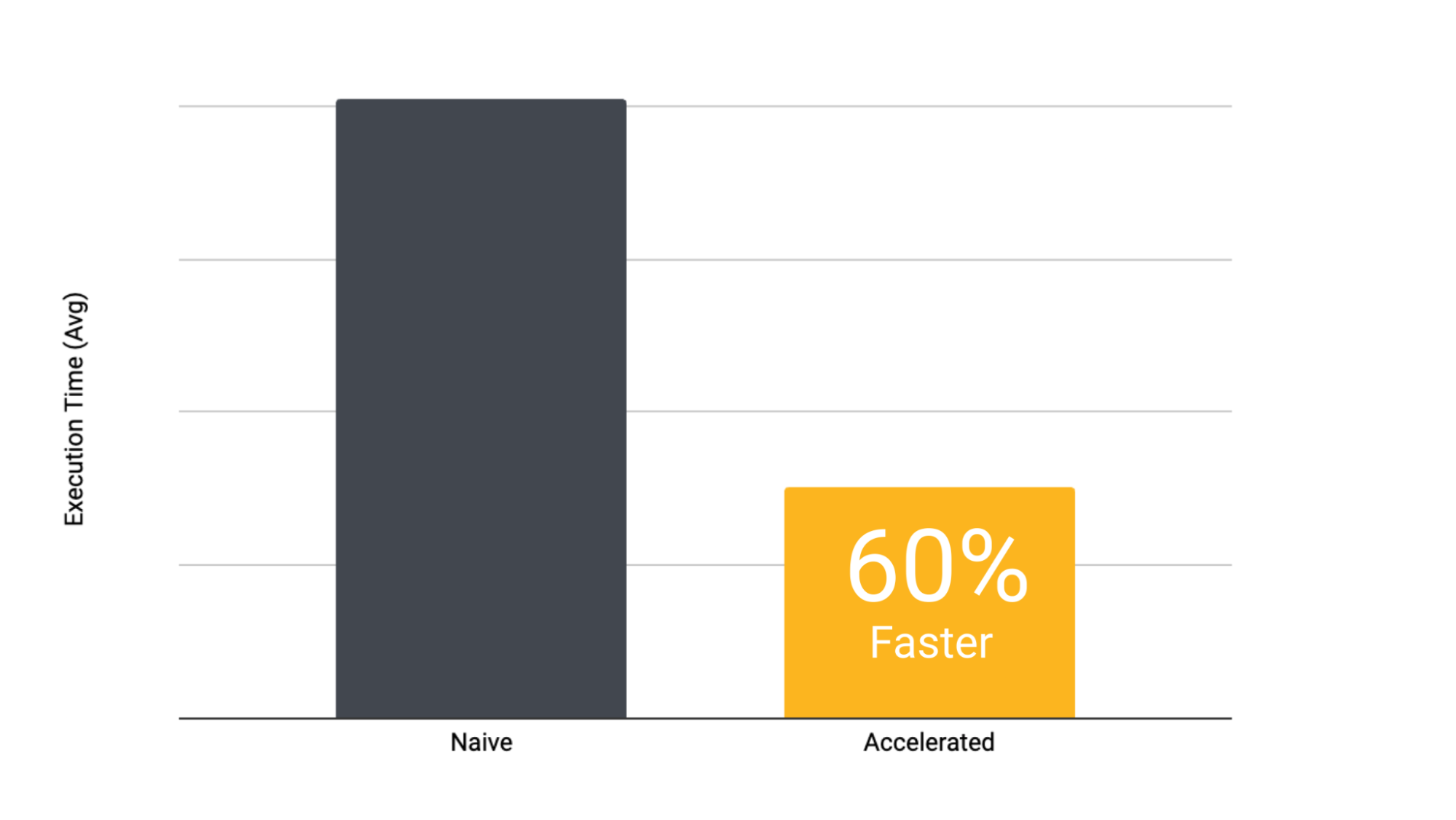
The total runtime is significantly longer for the non-accelerated version. There’s also some overhead in the first task due to the node being spun up.
Histograms plotting the runtimes in seconds for a task to complete (x-axis) vs. the number of executions (y-axis) shows that tasks using accelerated datasets are much faster than regular tasks. Capturing runtime within the task code controls for task scheduling and teardown overhead inherent in all executions.
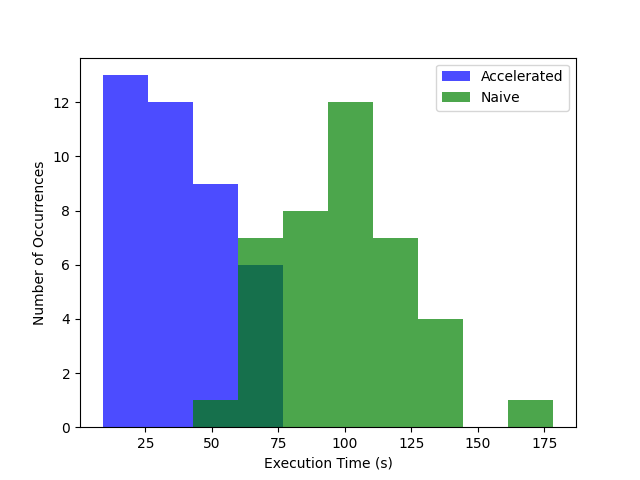
We attribute the consistently shorter runtimes for the tasks using accelerated datasets to the fact that the checksumming function could run asynchronously while chunks were fetched. It also appears that the s3-mountpoint is more performant than a naive S3 fetch, as all the task executions that didn’t use accelerated datasets are significantly longer. The longer fetch time, combined with a lack of caching and having to download the data prior to checksumming them, leads to a fairly significant runtime increase across the board.
Real-world impact: decrease time-to-solution
The above benchmarks are synthetic. Real-world performance is much more salient and the team at DelveBio has realized much more impactful speedups.
We were trying to tackle a problem with massive genomic index files that needed to be downloaded into every pod for our primary alignment task. This was a huge bottleneck. Before leveraging the accelerated datasets solution, we were opting to build the index on the fly (downloading the source data and building the minhash table from it) to minimize the data transfer at the expense of needing tons of RAM for each pod. With the persistent storage option, we are able to store the pre-built indices and also reduce the RAM requirement for each worker. The gains reduced the task execution time by roughly 50% and the RAM requirements by 75%, effectively quadrupling our throughput on the same node pool. —Brian O’Donovan, Sr Director of Bioinformatics & Computational Biology @ DelveBio
Delve’s offering empowers clinicians to identify neuro-infectious pathogens in cerebrospinal fluid so patients can get the right course of treatment as soon as possible; we’re thrilled to help them realize this mission. The accelerated dataset capability enables Delve to reduce time-to-solution while keeping compute costs low.
What’s Next?
We’ll continue to develop this feature in partnership with our customers to provide for a more seamless developer experience. This will eventually get rolled into Artifacts where you’ll be able to pick and choose which ones you want accelerated from the UI. We’re planning to bring this feature to GCP and also looking to expand it on AWS and GCP to enable reads and writes instead of the current read-only implementation. This will enable a fast and cost-effective storage space for sharing data between tasks and workflows.
This capability is available on Union’s BYOC tier. If you want to speed up your workflows, check out the docs and schedule a free demo with Union!

